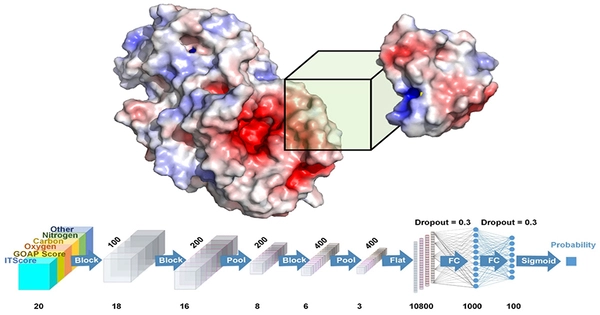
Indeed, neural networks have played an important role in protein design and computational biology. Protein design is a difficult challenge that entails predicting the amino acid sequences that will fold into the appropriate three-dimensional structure and perform particular functions.
To better comprehend and build proteins, a scientist blends attention neural networks and graph neural networks. The method combines the strengths of geometric deep learning and language models to predict known protein properties and imagine novel proteins that nature has not yet created. The model converts numbers, descriptions, tasks, and other information into symbols that can be used by neural networks.
Proteins, with their complicated arrangements and dynamic capabilities, execute a wide range of biological tasks by utilizing unique arrangements of simple building elements where geometry is critical. Researchers may be able to construct unique proteins for specific applications by translating this practically endless library of arrangements into their corresponding functions.
Markus Buehler of the Massachusetts Institute of Technology integrated attention neural networks, sometimes known as transformers, with graph neural networks to better understand and design proteins in the Journal of Applied Physics from AIP Publishing. The method combines the strengths of geometric deep learning and language models to not only predict existing protein features but also to imagine novel proteins that nature has not yet created.
While our current focus is on proteins, this method has vast potential in materials science. We’re particularly interested in investigating material failure behaviors, with the goal of designing materials with specific failure patterns.
Markus Buehler
“With this new method, we can utilize all that nature has invented as a knowledge basis by modeling the underlying principles,” Buehler said. “The model recombines these natural building blocks to achieve new functions and solve these types of tasks.”
Proteins have been notoriously difficult to simulate due to their complicated architectures, capacity to multitask, and tendency to alter shape when dissolved. Machine learning has shown the ability to transform the nanoscale dynamics that govern protein activity into functional frameworks. However, the reverse process of converting a desired function into a protein structure remains difficult.
Buehler’s technique overcomes this difficulty by converting numbers, descriptions, tasks, and other components into symbols that his neural networks can use. He began by training his model to predict the sequence, solubility, and amino acid building blocks of various proteins based on their functions. He then taught it to be inventive and generate entirely new structures in response to starting parameters for a new protein’s function.
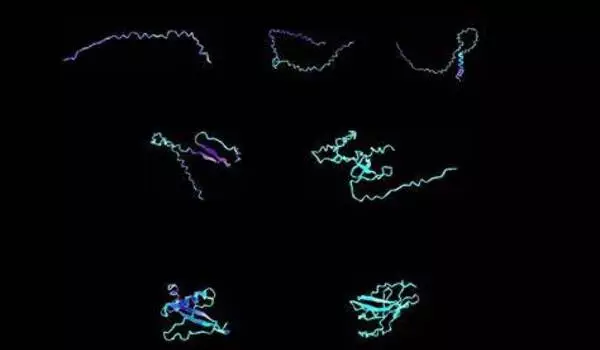
The approach allowed him to create solid versions of antimicrobial proteins that previously had to be dissolved in water. In another example, his team took a naturally occurring silk protein and evolved it into various new forms, including giving it a helix shape for more elasticity or a pleated structure for additional toughness.
The model performed many of the central tasks of designing new proteins, but Buehler said the approach can incorporate even more inputs for more tasks, potentially making it even more powerful.
“A big surprise was that the model performed exceptionally well despite being designed to solve multiple tasks.” This is most likely because the model learns more by examining a variety of tasks,” he explained. “Instead of developing specialized models for specific tasks, researchers can now think broadly in terms of multitask and multimodal models.”
Because of the comprehensive nature of this technique, this model can be used in many fields other than protein creation.
“While our current focus is on proteins, this method has vast potential in materials science,” Buehler added. “We’re particularly interested in investigating material failure behaviors, with the goal of designing materials with specific failure patterns.”